Is it possible obtain the sequence of a chloroplast genome from a RNA-Seq? Yes, it is. In this paper, we use a software pipeline to produce quality chloroplast genomes from RNA and compare it with genomic DNA-derived cp genomes. 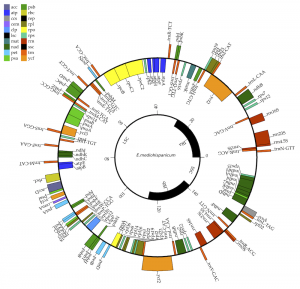
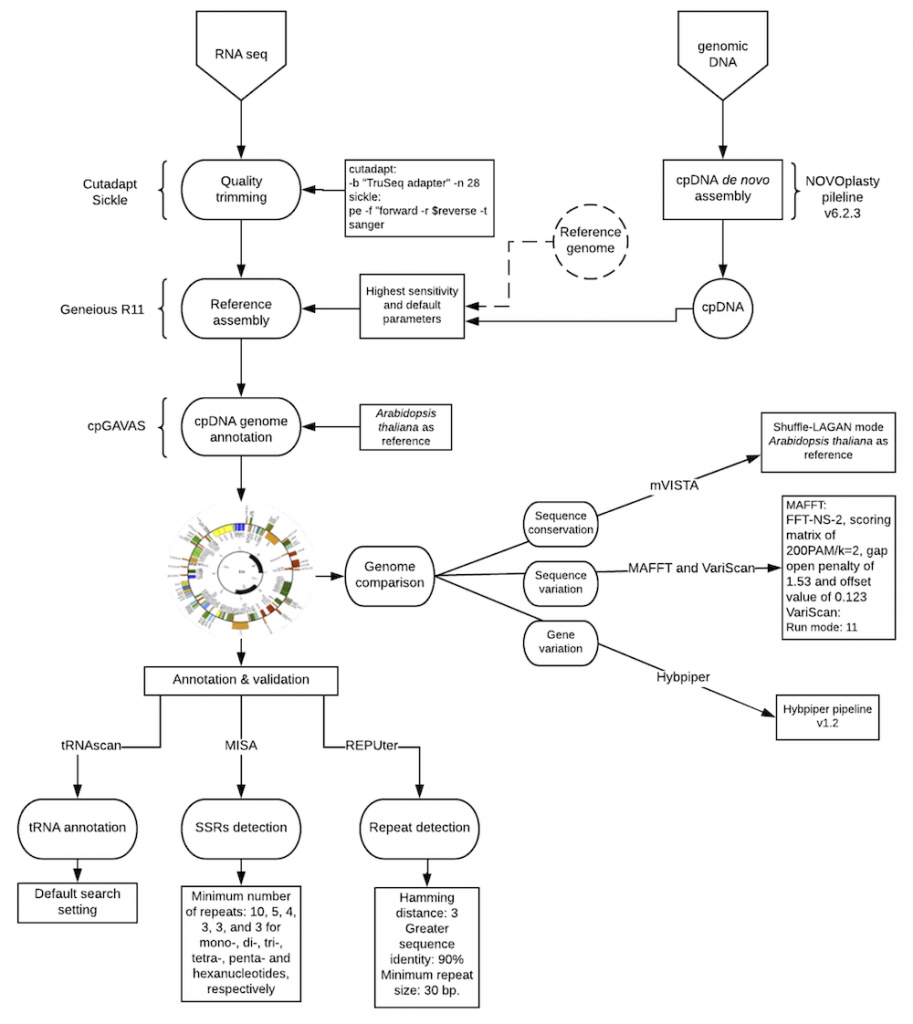
Chloroplast genomes (cp genomes) are widely used in comparative genomics, population genetics, and phylogenetic studies. Obtaining chloroplast genomes from RNA-Seq data seems feasible due to the almost full transcription of cpDNA. However, the reliability of chloroplast genomes assembled from RNA-Seq instead of genomic DNA libraries remains to be thoroughly verified. In this study, we assembled chloroplast genomes for three Erysimum (Brassicaceae) species from three RNA-Seq replicas and from one genomic library of each species, using a streamlined bioinformatics protocol. We compared these assembled genomes, confirming that assembled cp genomes from RNA-Seq data were highly similar to each other and to those from genomic libraries in terms of overall structure, size, and composition. Although post-transcriptional modifications, such as RNA-editing, may introduce variations in the RNA-seq data, the assembly of cp genomes from RNA-seq appeared to be reliable. Moreover, RNA-Seq assembly was less sensitive to sources of error such as the recovery of nuclear plastid DNAs (NUPTs). Although some precautions should be taken when producing reference genomes in non-model plants, we conclude that assembling cp genomes from RNA-Seq data is a fast, accurate, and reliable strategy.
More info and PDF:
Osuna-Mascaró C, Rubio de Casas R, Perfectti F. 2018.
Comparative assessment shows the reliability of chloroplast genome assembly using RNA-seq
Scientific Reports 8: 17404
https://doi.org/10.1038/s41598-018-35654-3
